The Protein Folding Answer
I need to issue a disclaimer before anything else. The script below is most definitely NOT the answer to protein folding. In fact, I make no claims at all about the efficacy of this code beyond it’s rudimentary ability to fold the beta-hairpin shown here. This code is just a homework assignment and learning experience for me. I believe the code is decently well commented for anyone who’s working with PyRosetta themselves. MAYBE this can be used at a starting point.
I’m also not going to go through the code section by section as I did for the sequence similarity code, but here’s a brief discussion of what the script is supposed to accomplish:
- First the peptide has to be generated. It’s what we are going to fold in this program.
- Since we know we’re making a beta-hairpin, we use knowledge based design to set the basic starting point angles of the backbone.
- Then we go through a million or so cycles of what amounts to three steps:
i. Randomly shift the peptide by some amount.
ii. Calculate the energy score of the new position compared to the old one. (Since we know we’re looking for hydrogen bonds, we add an extra bonus for moves that potentially form h-bonds)
iii. Decide whether to accept the new position of the peptide, or to reject it and keep the old position. This decision is based on the Metropolis Criteria. - As the folding simulation progresses we periodically output the energy score to see how things are going.
- At the end of the run, we save the new peptide so we can view it later.
In a way, the steps above are deceptively simply. There are only a few processes and they’re fairly straightforward. On the other hand, the theory underlying these steps are very powerful. More importantly, the amount of refinement that can be build on top of these steps is enormous.
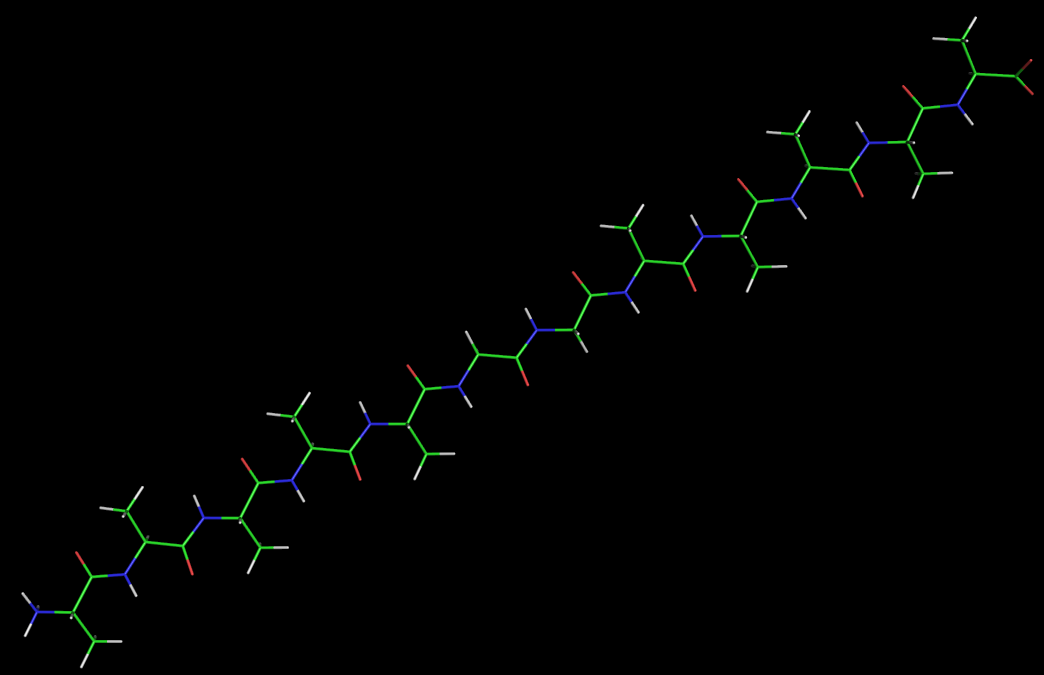
At the beginning of the simulation, the peptide is just a straight chain:
"""Import necessary libraries and initialize rosetta"""
import random
import math
from rosetta import *
from rosetta.PyMolLink import *
init()
"""Generate the beta-hairpin pose and score it"""
strand = "AAAAA" #alanine end residues
turn = "GG" #glycine turn residues
hairpin = strand+turn+strand #combine to make hairpin
pose = pose_from_sequence(hairpin,"fa_standard") #Create Rosetta pose
fa = get_fa_scorefxn() #Initialize a scoring function
fa(pose) #Score the hairpin
pose.dump_pdb("start1.pdb") #Generate an image of the starting structure
pose.set_psi(5,90) #Set starting angle
pose.set_phi(6,60) #Set starting angle
pose.set_psi(6,-120) #Set starting angle
pose.set_phi(7,-80) #Set starting angle
pose.set_psi(7,0) #Set starting angle
pose.set_phi(8,-120) #Set starting angle
"""Initialize variables"""
old_pose = Pose() #Initialize empty poses for later
low_pose = Pose() #Initialize empty poses for later
max_temp = 100 #Initial temperature for metropolis and annealing
current_temp = max_temp #Current temperature will drop through cycles
max_cycles = 1000000 #How many moves you want to make
low_energy = 10000 #Will store the value of lowest energy conformation
constraint_weight = 0.5 #Parameter for how much h-bond distances matter
ideal_dist = 0.29 #The proper distance for an h-bond between the alanines
"""Begin iterating through moves"""
for i in range(0,max_cycles): #Loop through the all the cycles
"""Update accept flag, and the old pose/energy/score"""
old_pose.assign(pose) #Set the old_pose
fa(old_pose) #Score the old pose
old_energy = fa.score(old_pose) #Define variable for the energy of old pose
old_score = old_energy #Alias old_energy, to use old_score for hbond constraining
"""Move given angle or angles by a chosen amount."""
current_residue = random.randint(1,pose.total_residue()) #Choose the residue to move from a uniform distribution
curr_phi = pose.phi(current_residue) #Set the current phi angle to be moved
curr_psi = pose.psi(current_residue) #Set the current psi angle to be moved
angle_shift = random.gauss(0,25*(float(i)/float(max_cycles))) #As the folding progresses, move the angle(s) by a smaller amount
if(random.random()<(float(i)/float(max_cycles))): #As the folding progresses, make the probability of a shear move increase
pose.set_phi(current_residue,curr_phi+angle_shift) #Set the new phi angle (shear move)
pose.set_psi(current_residue,curr_psi-angle_shift) #Set the new psi angle (shear move)
elif(random.random()<0 data-blogger-escaped--0.01="" data-blogger-escaped--="" data-blogger-escaped-.5="" data-blogger-escaped-0:="" data-blogger-escaped-0="" data-blogger-escaped-1000="" data-blogger-escaped-3="" data-blogger-escaped-a="" data-blogger-escaped-above="" data-blogger-escaped-accept:="" data-blogger-escaped-accept="" data-blogger-escaped-accepted="" data-blogger-escaped-accepting="" data-blogger-escaped-actions="" data-blogger-escaped-addition="" data-blogger-escaped-after="" data-blogger-escaped-alculate="" data-blogger-escaped-and="" data-blogger-escaped-angle="" data-blogger-escaped-angle_shift="" data-blogger-escaped-appropriate="" data-blogger-escaped-are="" data-blogger-escaped-as="" data-blogger-escaped-atom="" data-blogger-escaped-atoms="" data-blogger-escaped-ave="" data-blogger-escaped-bad="" data-blogger-escaped-between="" data-blogger-escaped-bonding="" data-blogger-escaped-change="" data-blogger-escaped-chosen="" data-blogger-escaped-code="" data-blogger-escaped-constraining="" data-blogger-escaped-constraint_weight="" data-blogger-escaped-core="" data-blogger-escaped-criteria="" data-blogger-escaped-cterm="pose.residue(12-2*k)" data-blogger-escaped-cterm_pos="Cterm.xyz(" data-blogger-escaped-curr_phi="" data-blogger-escaped-curr_psi="" data-blogger-escaped-current="" data-blogger-escaped-current_residue="" data-blogger-escaped-current_temp="max_temp*(math.exp(-1*max_cycles**-1))" data-blogger-escaped-delta_e="" data-blogger-escaped-dimentions="" data-blogger-escaped-dist="dist" data-blogger-escaped-distance="" data-blogger-escaped-each="" data-blogger-escaped-ecide="" data-blogger-escaped-edo="" data-blogger-escaped-efine="" data-blogger-escaped-else:="" data-blogger-escaped-end="" data-blogger-escaped-energy:="" data-blogger-escaped-energy="" data-blogger-escaped-epeat="" data-blogger-escaped-eset="" data-blogger-escaped-estore="" data-blogger-escaped-et="" data-blogger-escaped-f="" data-blogger-escaped-fa="" data-blogger-escaped-file="" data-blogger-escaped-final="" data-blogger-escaped-find="" data-blogger-escaped-first="" data-blogger-escaped-folding="" data-blogger-escaped-for="" data-blogger-escaped-hbond="" data-blogger-escaped-hbonding="" data-blogger-escaped-heck="" data-blogger-escaped-here="" data-blogger-escaped-i="" data-blogger-escaped-ideal_dist-old_dist="" data-blogger-escaped-if="" data-blogger-escaped-in="" data-blogger-escaped-inal="" data-blogger-escaped-ind="" data-blogger-escaped-is="" data-blogger-escaped-isn="" data-blogger-escaped-iterations="" data-blogger-escaped-j="" data-blogger-escaped-just="" data-blogger-escaped-k="" data-blogger-escaped-lias="" data-blogger-escaped-low_energy="" data-blogger-escaped-low_pose.assign="" data-blogger-escaped-low_pose.dump_pdb="" data-blogger-escaped-lower="" data-blogger-escaped-lowest="" data-blogger-escaped-math.fabs="" data-blogger-escaped-math.pow="" data-blogger-escaped-max_cycles="" data-blogger-escaped-metropolis="" data-blogger-escaped-move="" data-blogger-escaped-moves="" data-blogger-escaped-new="" data-blogger-escaped-new_dist="math.sqrt(dist)" data-blogger-escaped-new_energy="" data-blogger-escaped-new_score="new_score+math.fabs(ideal_dist-new_dist)*constraint_weight" data-blogger-escaped-not="" data-blogger-escaped-nterm="pose.residue(1+2*k)" data-blogger-escaped-nterm_pos="Nterm.xyz(" data-blogger-escaped-of="" data-blogger-escaped-old="" data-blogger-escaped-old_dist="math.sqrt(dist)" data-blogger-escaped-old_energy="" data-blogger-escaped-old_pose="" data-blogger-escaped-old_score="" data-blogger-escaped-one="" data-blogger-escaped-onte_carlo_2-blog2.pdb="" data-blogger-escaped-or="" data-blogger-escaped-other="" data-blogger-escaped-outcome.="" data-blogger-escaped-pair="" data-blogger-escaped-pdate="" data-blogger-escaped-pdb="" data-blogger-escaped-phi="" data-blogger-escaped-pose.="" data-blogger-escaped-pose.assign="" data-blogger-escaped-pose.set_phi="" data-blogger-escaped-pose.set_psi="" data-blogger-escaped-pose="" data-blogger-escaped-poses="" data-blogger-escaped-pre="" data-blogger-escaped-print="" data-blogger-escaped-probability="" data-blogger-escaped-process="" data-blogger-escaped-progress="" data-blogger-escaped-psi="" data-blogger-escaped-r="" data-blogger-escaped-range="" data-blogger-escaped-residue="" data-blogger-escaped-residues="" data-blogger-escaped-rint="" data-blogger-escaped-runs="" data-blogger-escaped-save="" data-blogger-escaped-score.="" data-blogger-escaped-score="" data-blogger-escaped-script="" data-blogger-escaped-second="" data-blogger-escaped-set="" data-blogger-escaped-shear="" data-blogger-escaped-so="" data-blogger-escaped-spacial="" data-blogger-escaped-statements="" data-blogger-escaped-str="" data-blogger-escaped-structure="" data-blogger-escaped-t="" data-blogger-escaped-take="" data-blogger-escaped-temperature="" data-blogger-escaped-term_pos="" data-blogger-escaped-than="" data-blogger-escaped-the="" data-blogger-escaped-there="" data-blogger-escaped-to="" data-blogger-escaped-until="" data-blogger-escaped-update="" data-blogger-escaped-use="" data-blogger-escaped-utoff="" data-blogger-escaped-value="" data-blogger-escaped-values="" data-blogger-escaped-variable="" data-blogger-escaped-very="" data-blogger-escaped-whether="" data-blogger-escaped-with="">